- Institute
- Research topics
- Organization
- Platforms
- Services
- Europe/International
- Science outreach
- Agenda
- Directory
- Access
Heterochromatin and Polycomb-marked gene Silencing by RNA decay
We have recently discovered that the LSM2-8 complex mediates a mechanism of RNA degradation that selectively targets H3K27me3-marked genes in  C. elegans, revealing that in animals heterochromatin can be silenced by specific degradation of heterochromatic transcripts, and not only by transcriptional repression. Therefore, we are interested to focus our research on post-transcriptional mechanism(s) involved in heterochromatin silencing in general in higher eukaryotes. We expect that this research will deepen our understanding of regulation of gene expression in normal animal development and in response to the environment by integrating this novel RNA silencing regulation. In a longer perspective, our goal is to better understand the crosstalk between RNA and chromatin implicated in gene regulation, the initial targeting of the Polycomb H3K27me3 mark to the genome in C. elegans and mammals and ultimately to inflect abnormal gene expression in pathological contexts.
C. elegans, revealing that in animals heterochromatin can be silenced by specific degradation of heterochromatic transcripts, and not only by transcriptional repression. Therefore, we are interested to focus our research on post-transcriptional mechanism(s) involved in heterochromatin silencing in general in higher eukaryotes. We expect that this research will deepen our understanding of regulation of gene expression in normal animal development and in response to the environment by integrating this novel RNA silencing regulation. In a longer perspective, our goal is to better understand the crosstalk between RNA and chromatin implicated in gene regulation, the initial targeting of the Polycomb H3K27me3 mark to the genome in C. elegans and mammals and ultimately to inflect abnormal gene expression in pathological contexts.
All our cells have the same genome, with the same genes, but different combinations of genes are expressed or repressed/silenced to create different cell types and to adapt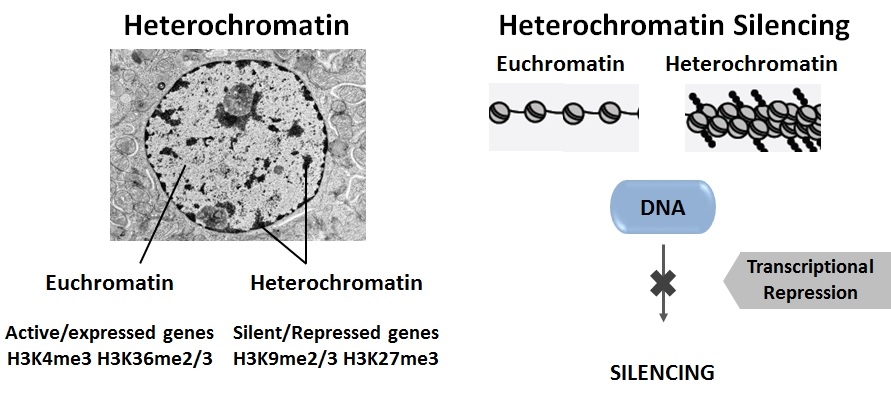 to the environment. The expressed genes (DNA) are transcribed into RNA molecules and, in most cases, translated into proteins. The regulation of the expression of genes into RNA and proteins is essential for life.
to the environment. The expressed genes (DNA) are transcribed into RNA molecules and, in most cases, translated into proteins. The regulation of the expression of genes into RNA and proteins is essential for life.
The formation of heterochromatin, which is the condensed compartment of the genome, often accumulates as cells differentiate, and correlates with silencing of the expression of genes found in it. A chemical mark called “H3K27me3” or the “Polycomb mark” is found on some of the heterochromatin regions and this mark plays a major role in decisions about cell differentiation and maintenance of cellular identity. Misregulation of H3K27me3 levels contributes to many types of cancer (leukemia, lymphoma and others), as well as to genetic diseases.
Until our recent publication, silencing of genes associated with H3K27me3 had been explained exclusively by transcriptional repression which implies that those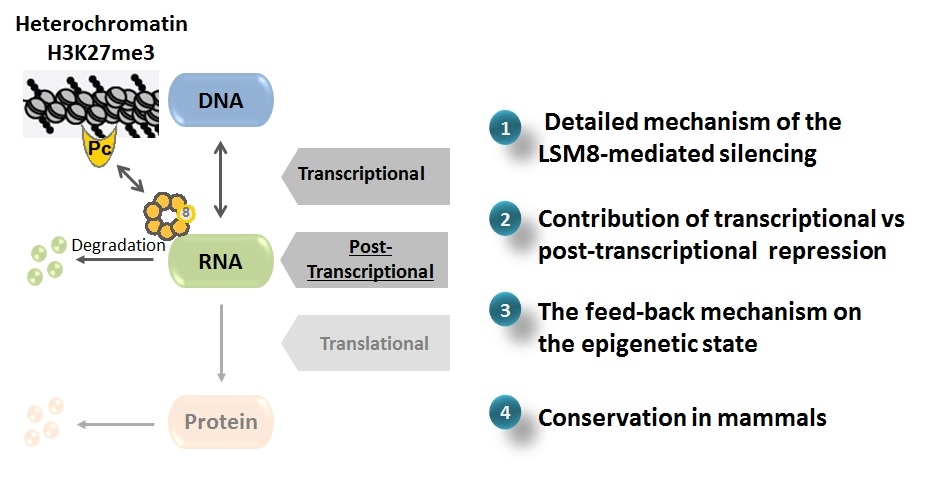 genes/DNA sequences are not transcribed into RNA. However, we have discovered that many H3K27me3-marked genes are actually transcribed and that there is another type of regulation at the post-transcriptional level that specifically degrades RNA arising from those genes, enriched in H3K27me3. The RNA binding complex “LSM2-8” and the exoribonuclease “XRN-2” are involved in this process.
genes/DNA sequences are not transcribed into RNA. However, we have discovered that many H3K27me3-marked genes are actually transcribed and that there is another type of regulation at the post-transcriptional level that specifically degrades RNA arising from those genes, enriched in H3K27me3. The RNA binding complex “LSM2-8” and the exoribonuclease “XRN-2” are involved in this process.
We will focus our research on this novel mechanism of heterochromatic RNA degradation, which will allow a better understanding of this post-transcrptional regulation and its potential contribution to cancer. In a longer term, this could be crucial for the development of innovative therapeutic targeting.
I have been recently recruited at the CBI as a Junior Group leader and got a CRCN (INSERM) position. Therefore, if you are interested to discuss Science or to join my emerging group, please do not hesitate to contact me directly!
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Dr. Anna MATTOUT
anna.mattout@univ-tlse3.fr
INSERM, CRCN,
Centre de Biologie Integrative (CBI)
169 Avenue Grunberg-Manago, MCD-UMR 5077
4R4 Building, 3rd floor, Lab 3144, Office 3045
31062 Toulouse, France
+33(0)5 61 55 87 07; +33(0)6 44 74 26 56
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~






