- Institute
- Research topics
- Organization
- Platforms
- Services
- Europe/International
- Science outreach
- Agenda
- Directory
- Access

For more information on our research and our team, please visit our website: https://www.legubelab.com/
Come and visit Raphaël Mourad's webpage here.
Projects
Among the types of damage that can challenge the DNA molecule, DNA Double Strands Breaks (DSBs) are the most deleterious since they can lead to various mutations and chromosomes rearrangements. This type of damage can arise during development as part of programmed processes (during immune system formation for example) but also due to various environmental stress (cigarette smoke, irradiation,..). Over the past few years it has become evident that chromatin, that packages DNA in eukaryotes nuclei and that is encoding epigenetic information, is the real substrate for all DNA related processes and plays a decisive role in DNA repair. Repair of DSB into the chromatin context raises several questions that we aim to address in the lab.
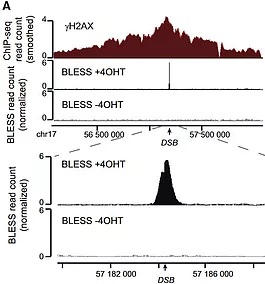
To address these questions, we developed the DIvA cell line which is a powerful experimental cell system that enables the creation of DSBs at well known positions across the genome in various chromatin contexts and which has the advantage of detaining statistical power over a large range of DSBs
DIvA cells based on the expression of a restriction enzyme fused to a modified estrogen receptor ligand binding domain and to an auxin-inducible degron (AID-AsiSI-ER). 4OHT treatment of this cell line triggers the nuclear localization of the AsiSI restriction enzyme and the rapid induction (<1 hour) of multiples (about 150) sequence-specific DSBs, widespread across the genome (Iacovoni et al, 2010; Massip et al, 2010). An additional auxin treatment leads to the rapid degradation of the enzyme thus allowing repair completion (Aymard et al, 2014). In other words this system allows a “pulse-chase” induction of DSB at annotated sites, dispersed on the human genome.

How does the chromatin state influence the recognition of DSB and the repair pathway choice?
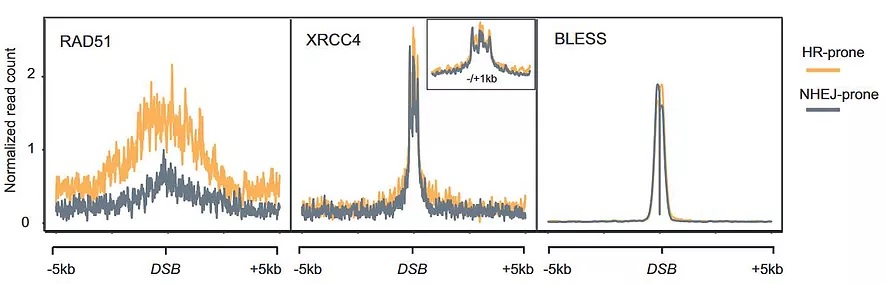
In eukaryotes, DSB repair occurs within chromatin which is the structure that tightly packages and regulates DNA metabolism. A fair amount of studies, including ours, have suggested a role of chromatin in addressing the repair pathway. Our hypothesis is that a "repair histone code" exists, where specific histone marks present before the appearance of a DSB, could specifically help to recruit and/or stabilize one or the other repair pathway (Clouaire, et al, 2015). And indeed, chromatin influences DSB repair at various steps, from the break detection to sequence recovery, with for example the histone mark H3K36me3 being important for channeling DSB induced in active genes towards HR (Aymard et al, 2014). Importantly, chromatin can also be altered during the repair process, and this at various scales, in a manner that relates to the repair pathway used (Clouaire et al. 2018).
In order to decipher the chromatin landscape induced at DSBs, we mainly used BLESS, ChIP-chip/seq and RNA seq to draw high resolution profiles of DSB-induced chromatin modifications and DNA repair complexes around breaks (Iacovoni et al, 2010; Massip et al, 2010; Caron et al, 2012; Clouaire et al, 2018).
How are active genes repaired?
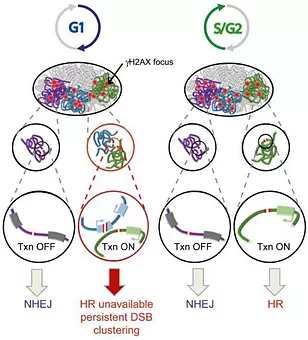
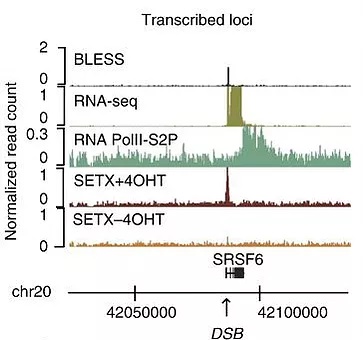
Recent work revealed that transcriptionally active loci are prone to breakage and our work suggested that when damaged, active genes display a very peculiar behavior compared to the rest of the genome: they experience a strong repair delay and undergo clustering in an actin/LINC and MRN-dependent manner in G1 (Aymard et al, 2017) while they are mostly repaired by homologous recombination in a Senataxin-dependent manner in post-replicative cells (Cohen et al, 2018). We are trying to characterize in more details this novel transcription-coupled DSB Repair (TC-DSBR) pathway (reviewed in Marnef et al, 2018).
How does chromosome conformation changes following DSB?
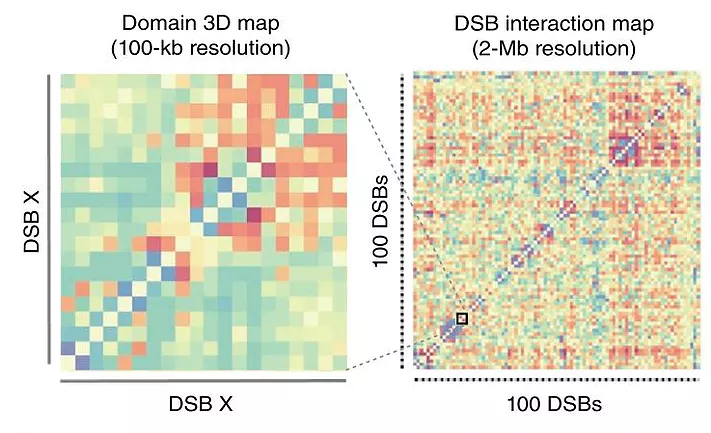
Our previous work deciphered the histone modifications landscape induced at DSBs. Yet, the structure of chromatin (nucleosome density, folding) in cis around DSB remains to be determined. Using chromosome conformation capture, we try to understand the structure in 3D of the chromosomes following breakage (such as (Aymard et al. 2017) where genome-wide mapping of long-range contacts using Capture-HiC enable us to unveil the clustering of DNA double-strand breaks at damaged active genes.

LEGUBE TEAM 2023












