- Institute
- Research topics
- Organization
- Platforms
- Services
- Europe/International
- Science outreach
- Agenda
- Directory
- Access
DiSilencing of transposable Elements (ETs)
Finely tuned gene transcription requires a dynamic regulation of chromatin structure, and the precise role of histone marks in this process is a subject of intense study. In particular, little is known about the role of chromatin marks in Transposable Element (TE) regulation. TEs are major components of the genome, constituting approximately 20% of the Drosophila genome and up to 50% in humans. TEs are genetic elements that threaten genomic stability through their capability to move throughout the genome. Strict regulation of TE expression appears essential for normal development but also for the normal physiology of somatic adult tissues, and their aberrant regulation is linked to infertility, cancer and neurodegenerative diseases. Understanding how transposable elements are regulated within the genome is thus a fundamental question in biology. Although it is known that the heterochromatin machinery is implicated in TE silencing, the mechanistic aspects of this regulation have not been established.
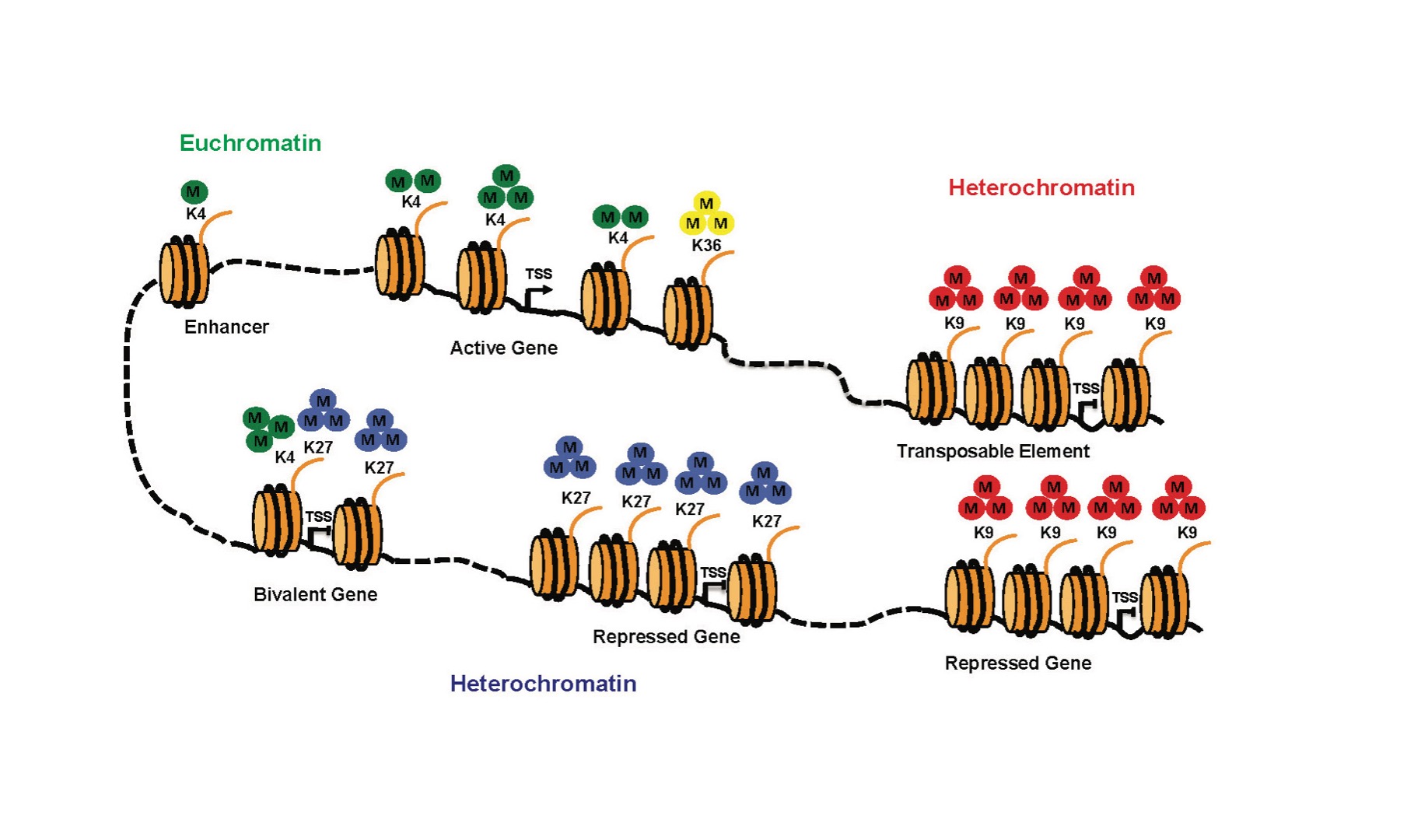
Di Stefano, Cells 2022
Our research focuses on the chromatin mediated mechanisms implicated in the control of gene and transposable element transcription through the study of chromatin modulation by the highly conserved histone demethylase, LSD1 and its co-factors. By combining genomic and proteomic approaches with genetic analysis and genome editing in the model organism Drosophila melanogaster we aim to:
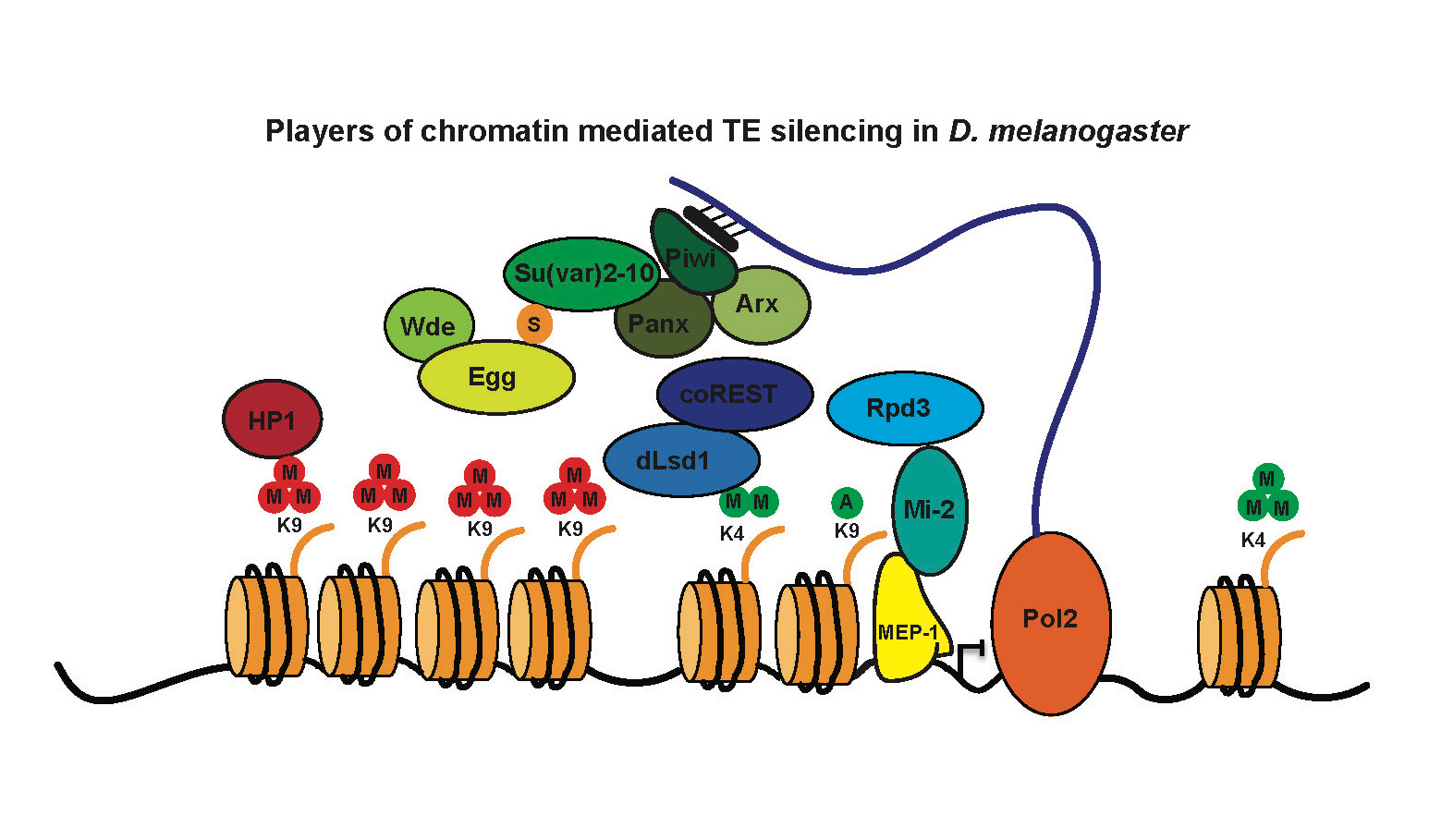
Di Stefano, Cells 2022



